GPT-4, Google Gemini Fall Short in Breast Imaging Classification
Released: April 30, 2024
- RSNA Media Relations
1-630-590-7762
media@rsna.org - Linda Brooks
1-630-590-7738
lbrooks@rsna.org - Imani Harris
1-630-481-1009
iharris@rsna.org
OAK BROOK, Ill. — Use of publicly available large language models (LLMs) resulted in changes in breast imaging reports classification that could have a negative effect on patient management, according to a new international study published today in the journal Radiology, a journal of the Radiological Society of North America (RSNA). The study findings underscore the need to regulate these LLMs in scenarios that require high-level medical reasoning, researchers said.
LLMs are a type of artificial intelligence (AI) widely used today for a variety of purposes. In radiology, LLMs have already been tested in a wide variety of clinical tasks, from processing radiology request forms to providing imaging recommendations and diagnosis support.
Publicly available generic LLMs like ChatGPT (GPT 3.5 and GPT-4) and Google Gemini (formerly Bard) have shown promising results in some tasks. Importantly, however, they are less successful at more complex tasks requiring a higher level of reasoning and deeper clinical knowledge, such as providing imaging recommendations. Users seeking medical advice may not always understand the limitations of these untrained programs.
"Evaluating the abilities of generic LLMs remains important as these tools are the most readily available and may unjustifiably be used by both patients and non-radiologist physicians seeking a second opinion," said study co-lead author Andrea Cozzi, M.D., Ph.D., radiology resident and post-doctoral research fellow at the Imaging Institute of Southern Switzerland, Ente Ospedaliero Cantonale, in Lugano, Switzerland.
Dr. Cozzi and colleagues set out to test the generic LLMs on a task that pertains to daily clinical routine but where the depth of medical reasoning is high and where the use of languages other than English would further stress LLMs capabilities. They focused on the agreement between human readers and LLMs for the assignment of Breast Imaging Reporting and Data System (BI-RADS) categories, a widely used system to describe and classify breast lesions.
The Swiss researchers partnered with an American team from Memorial Sloan Kettering Cancer Center in New York City and a Dutch team at the Netherlands Cancer Institute in Amsterdam.
The study included BI-RADS classifications of 2,400 breast imaging reports written in English, Italian and Dutch. Three LLMs—GPT-3.5, GPT-4 and Google Bard (now renamed Google Gemini)—assigned BI-RADS categories using only the findings described by the original radiologists. The researchers then compared the performance of the LLMs with that of board-certified breast radiologists.
The agreement for BI-RADS category assignments between human readers was almost perfect. However, the agreement between humans and the LLMs was only moderate. Most importantly, the researchers also observed a high percentage of discordant category assignments that would result in negative changes in patient management. This raises several concerns about the potential consequences of placing too much reliance on these widely available LLMs.
According to Dr. Cozzi, the results highlight the need for regulation of LLMs when there is a highly likely possibility that users may ask them health-care-related questions of varying depth and complexity.
"The results of this study add to the growing body of evidence that reminds us of the need to carefully understand and highlight the pros and cons of LLM use in health care," he said. "These programs can be a wonderful tool for many tasks but should be used wisely. Patients need to be aware of the intrinsic shortcomings of these tools, and that they may receive incomplete or even utterly wrong replies to complex questions."
The Swiss researchers were supervised by the co-senior author Simone Schiaffino, M.D. The American team was led by the co-first author Katja Pinker, M.D., Ph.D., and the Dutch team was led by the co-senior author Ritse M. Mann, M.D., Ph.D.
"BI-RADS Category Assignments by GPT-3.5, GPT-4, and Google Bard: A Multilanguage Study." Collaborating with Drs. Cozzi, Pinker, Schiaffino and Mann were Andri Hidber, B.Med., Tianyu Zhang, Ph.D., Luca Bonomo, M.D., Roberto Lo Gullo, M.D., Blake Christianson, M.D., Marco Curti, M.D., Stefania Rizzo, M.D., Ph.D., and Filippo Del Grande, M.D., M.B.A., M.H.E.M.
Radiology is edited by Linda Moy, M.D., New York University, New York, N.Y., and owned and published by the Radiological Society of North America, Inc. (https://pubs.rsna.org/journal/radiology)
RSNA is an association of radiologists, radiation oncologists, medical physicists and related scientists promoting excellence in patient care and health care delivery through education, research and technologic innovation. The Society is based in Oak Brook, Illinois. (RSNA.org)
For patient-friendly information on breast imaging, visit RadiologyInfo.org.
Images (JPG, TIF):
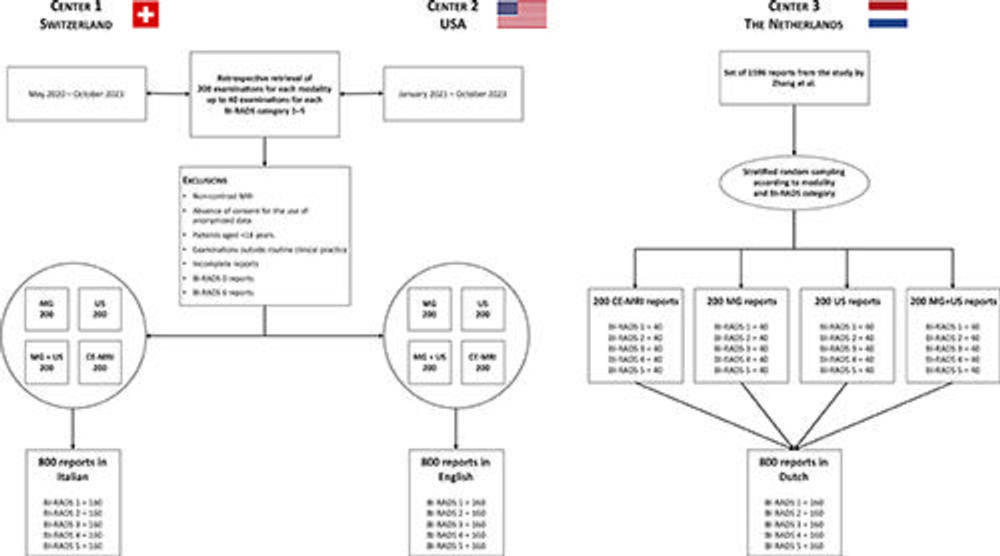
Figure 1. Study flowchart. BI-RADS = Breast Imaging Reporting and Data System, CE-MRI = contrast-enhanced MRI, MG = mammography.
High-res (TIF) version
(Right-click and Save As)
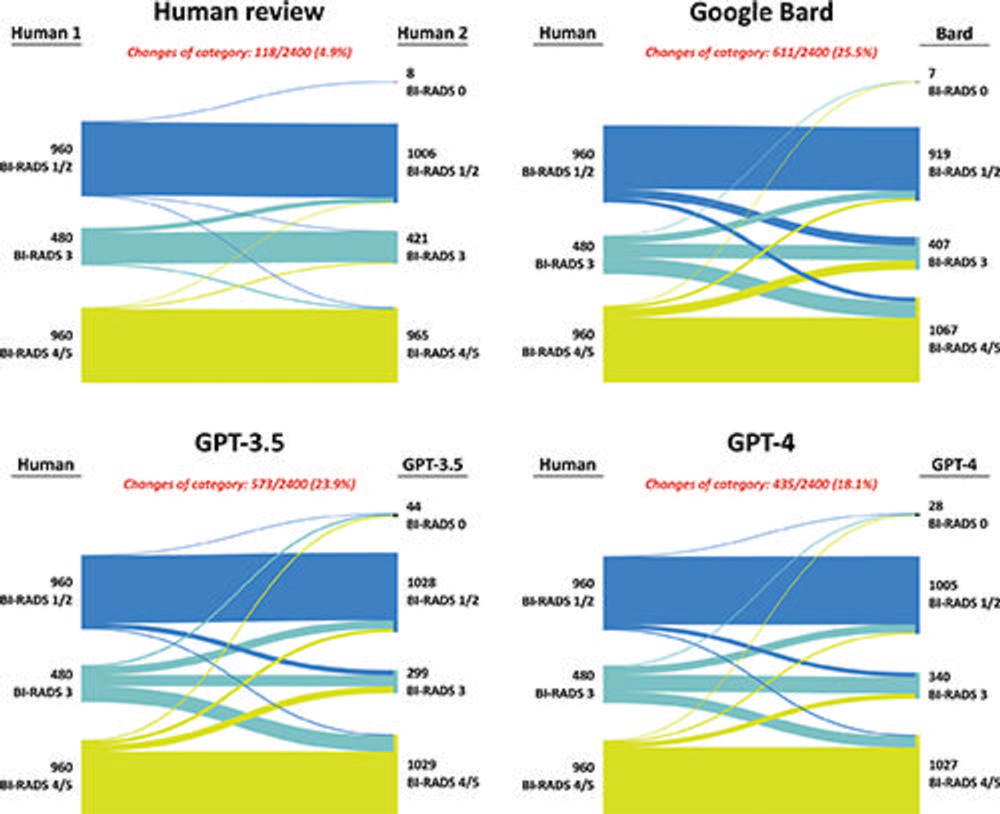
Figure 2. Sankey plots showing changes in Breast Imaging Reporting and Data System (BI-RADS) clinical management categories between human readers and between human readers and large language models (LLMs). Human-human agreement was assessed between the original radiologists who wrote the breast imaging reports (Human 1) and the radiologists who reviewed the findings section of the reports (Human 2). Human-LLM agreement was assessed between the original breast imaging reports and the outputs from three LLMs (Google Bard, GPT-3.5, and GPT-4) provided with the findings section of the report. The proportion of disagreements between the original reporting radiologists and the radiologists who reviewed the findings section of the reports was observed to be lower than the proportions of disagreements between the original reporting radiologists and the LLMs.
High-res (TIF) version
(Right-click and Save As)
