Machine Learning Method Identifies Precancerous Colon Polyps
Released: February 23, 2021
At A Glance
- A machine learning algorithm can predict the character of individual polyps based on quantitative image features extracted through radiomics.
- In the test set, the machine learning approach enabled noninvasive differentiation of benign and premalignant CT colonography-detected colorectal polyps.
- The findings point to a role for machine learning-derived algorithms in boosting the effectiveness of CT colonography as a screening tool for colorectal cancer.
- RSNA Media Relations
1-630-590-7762
media@rsna.org - Linda Brooks
1-630-590-7738
lbrooks@rsna.org - Katherine Anderson
1-630-491-1009
kanderson@rsna.org
OAK BROOK, Ill. — A machine learning algorithm helps accurately differentiate benign and premalignant colorectal polyps on CT colonography scans, according to a study published in the journal Radiology.
Colorectal cancer is among the three most common causes of cancer-related death among men and women in industrialized countries. Most types of colorectal cancer originate from adenomatous polyps—gland-like growths on the mucous membrane lining the large intestine—that develop over several years. Early detection and removal of these precancerous polyps can reduce the incidence and mortality of colorectal cancer.
During the last two decades, CT colonography emerged as a noninvasive alternative to colonoscopy in screening for colorectal cancer. It is comparable to colonoscopy in detecting most polyps and is effective at visualizing portions of the colon that in cases of complex anatomical conditions cannot always be evaluated by colonoscopy. However, CT colonography does not enable a definite differentiation between benign and premalignant polyps, which is crucial for individual risk stratification and therapy guidance.
For the new study, researchers leveraged the power of radiomics, a process of extracting quantitative features from medical images, to characterize polyps beyond what was apparent to the naked eye.
The researchers developed a machine learning algorithm to predict the character of the individual polyps based on quantitative image features extracted through radiomics. They applied the noninvasive, radiomics-based machine learning method on CT colonography images from a group of asymptomatic patients at average risk of colorectal cancer. The machine learning algorithm was trained on a set of more than 100 colorectal polyps in 63 patients and then tested on a set of 77 polyps in 59 patients.
In the test set, the machine learning approach enabled noninvasive differentiation of benign and premalignant CT colonography-detected colorectal polyps, with a sensitivity of 82%, and specificity of 85%. The area under the curve (AUC), a graphical measurement that reflects how much the model is capable of distinguishing between benign and precancerous polyps, was excellent.
"These results serve as proof-of-concept that machine learning-based image analysis allows the noninvasive differentiation of benign and premalignant colorectal polyps in CT colonography data sets," said study lead author Sergio Grosu, M.D., radiologist from University Hospital, Ludwig Maximilian University of Munich, in Munich, Germany. "The AUC of 0.91 indicates that this method works well."
The findings point to a role for machine learning-derived algorithms in boosting the effectiveness of CT colonography as a screening tool for colorectal cancer.
"Adding machine learning-assisted image analysis to conventional, radiological image reading could further improve the clinical significance of CT colonography-based colorectal cancer screening by allowing for a more precise selection of patients eligible for subsequent polypectomy," Dr. Grosu said. "This method could be used routinely as a second reader in all CT colonography examinations in the distant future."
Dr. Grosu said that additional studies with larger numbers of patients are needed to validate the findings. He added that these studies should also help drive improvements in the machine learning algorithm.
"Further refinement of the machine learning-based image analysis is necessary to achieve higher precision in polyp differentiation as well as workflow optimization for better applicability in clinical routine," Dr. Grosu said.
"Machine Learning-based Differentiation of Benign and Premalignant Colorectal Polyps Detected with CT Colonography in an Asymptomatic Screening Population – A Proof-of-Concept Study." Collaborating with Dr. Grosu were Philipp Wesp, M.Sc., Anno Graser, M.D., Stefan Maurus, M.D., Christian Schulz, M.D., Thomas Knösel, M.D., Clemens C. Cyran, M.D., Jens Ricke, M.D., Michael Ingrisch, Ph.D., and Philipp M. Kazmierczak, M.D.
Radiology is edited by David A. Bluemke, M.D., Ph.D., University of Wisconsin School of Medicine and Public Health, Madison, Wisconsin, and owned and published by the Radiological Society of North America, Inc. (https://pubs.rsna.org/journal/radiology)
RSNA is an association of radiologists, radiation oncologists, medical physicists and related scientists promoting excellence in patient care and health care delivery through education, research and technologic innovation. The Society is based in Oak Brook, Illinois. (RSNA.org)
For patient-friendly information on CT colonography, visit RadiologyInfo.org.
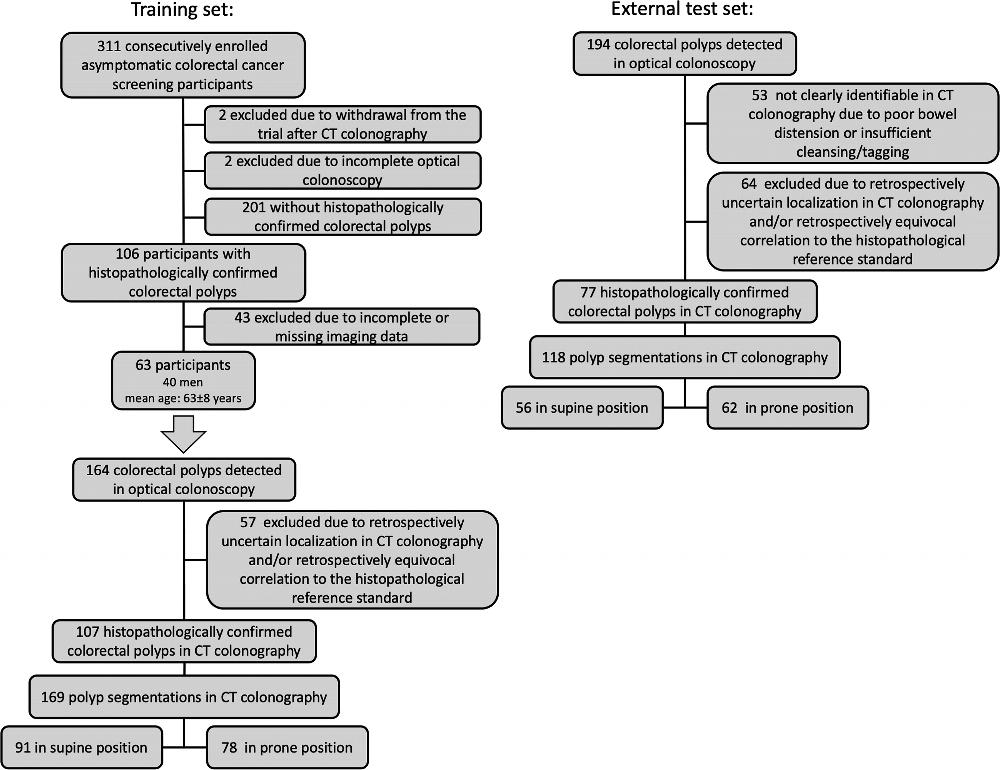
Figure 1. Flow diagram of the training set and external test set.
High-res (TIF) version
(Right-click and Save As)
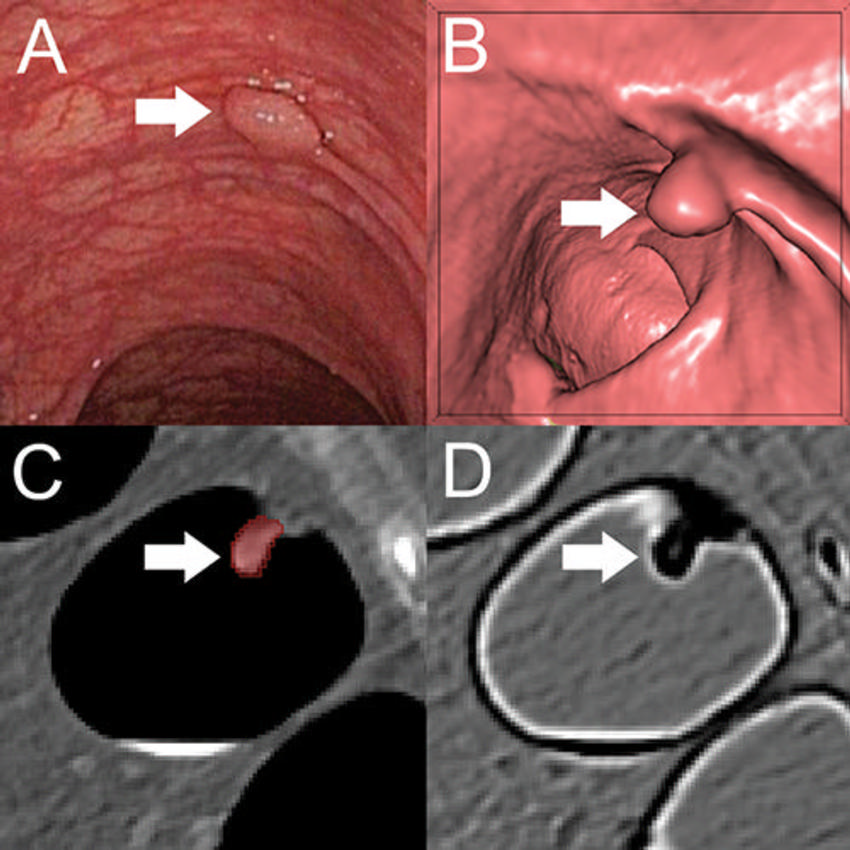
Figure 2. A, Optical colonoscopy and, B–D, CT colonography of a 9-mm polyp (arrow) in the descending colon of a 78-year-old woman. B, Virtual fly-through three-dimensional reconstructions were used for exact polyp localization. C, Manual polyp segmentation was performed in multiplanar two-dimensional CT colonography images. D, CT colonography images were preprocessed for image feature extraction by application of a dedicated filter.
High-res (TIF) version
(Right-click and Save As)
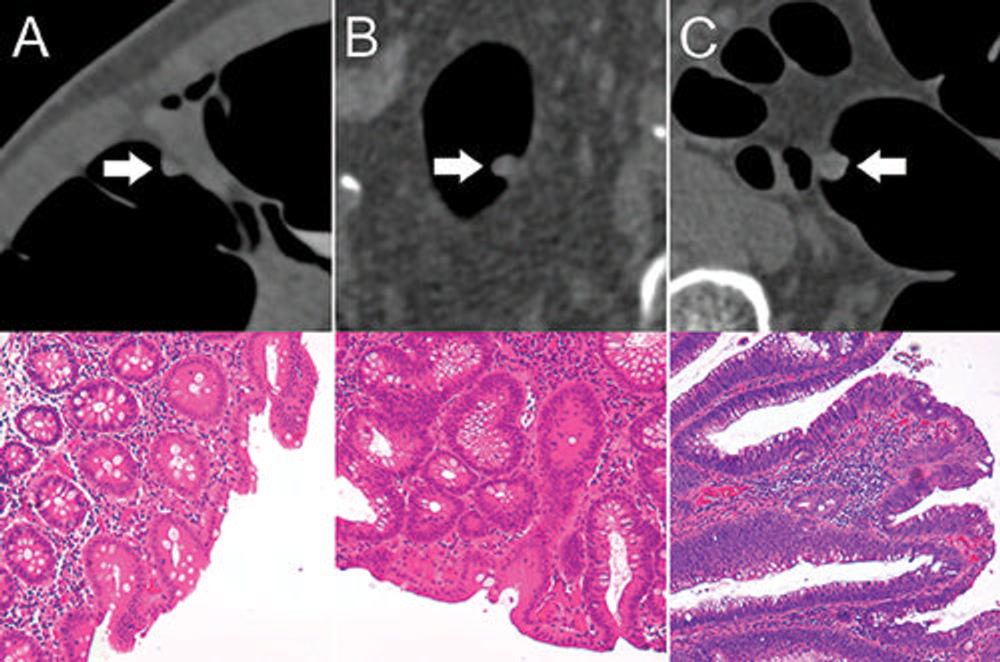
Figure 3. Top: Axial CT colonography images show representative colorectal polyps (arrow) in the training set. Bottom: Corresponding histopathologic work-up. (Hematoxylin-eosin staining; original magnification 320.) A, An 8-mm hyperplastic polyp in the ascending colon of a 54-year-old woman with hyperplastic epithelia. B, An 8-mm tubular adenoma in the sigmoid colon of a 68-year-old man with tubular growth pattern and elongated nuclei. C, An 11-mm tubulovillous adenoma in the rectum of a 73-year-old man with tubulovillous growth pattern and elongated nuclei.
High-res (TIF) version
(Right-click and Save As)
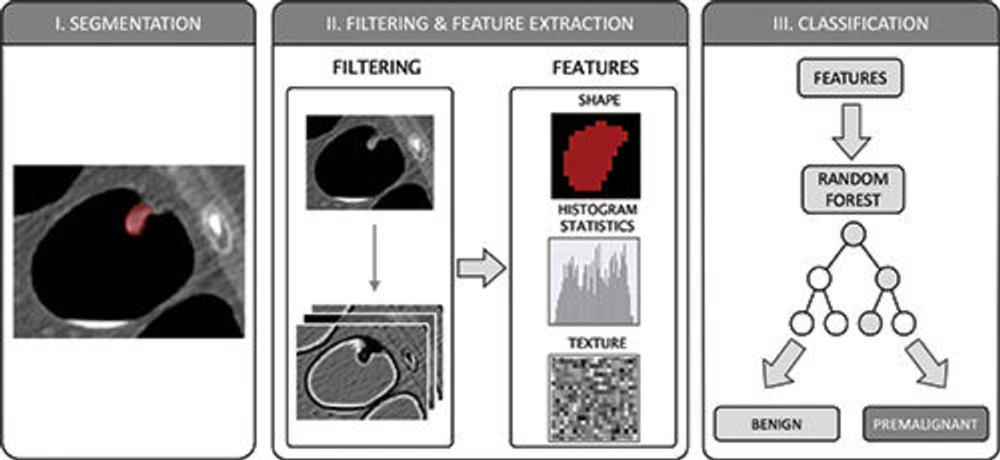
Figure 4. The radiomics workflow comprised three steps: manual segmentation of colorectal polyps in multiplanar two-dimensional CT colonography images; image filtering and feature extraction characterizing shape, histogram statistics, or texture; and feature-based training of a random forest classification algorithm to differentiate between benign and premalignant colorectal polyps according to the histopathologic reference standard.
High-res (TIF) version
(Right-click and Save As)
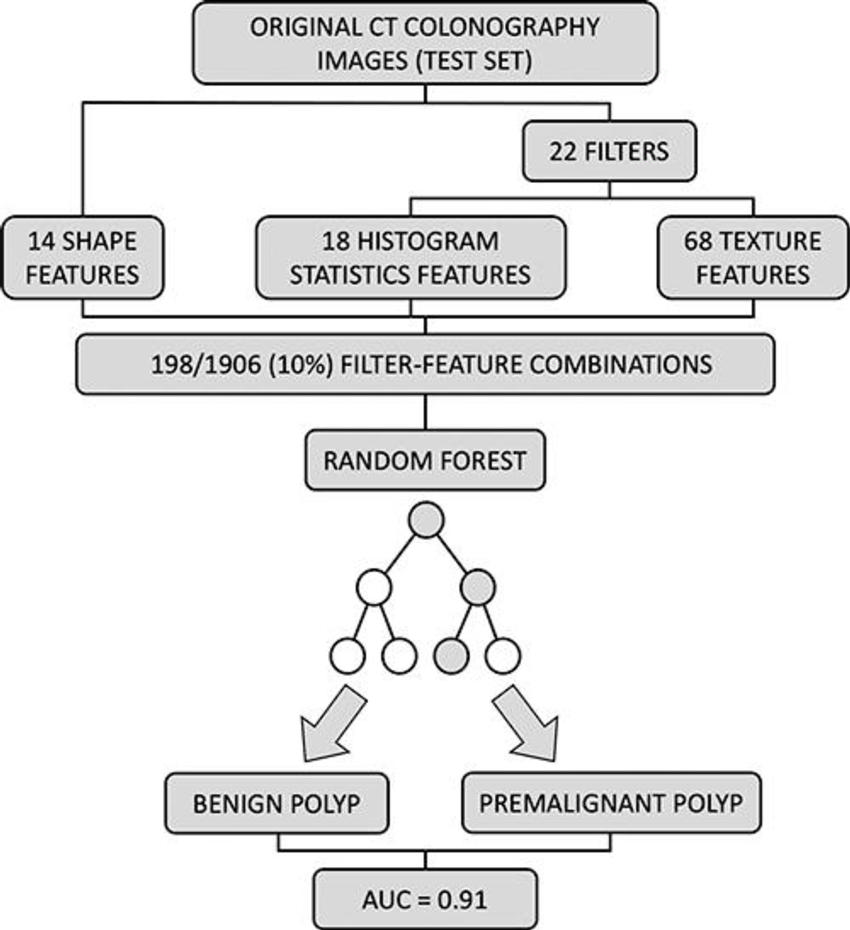
Figure 5. External validation of the trained random forest model. A total 198 of 1906 (10%) feature filter combinations were extracted from original CT colonography images of the test set using different image filters (n = 22) and image features characterizing shape (n = 14), histogram statistics (n = 18), or texture (n = 68). On the basis of these filter feature combinations, the trained random forest classifier was used to predict the colorectal polyp class label (benign vs premalignant). Prediction performance was quantified using area under the receiver operating characteristic curve (AUC).
High-res (TIF) version
(Right-click and Save As)
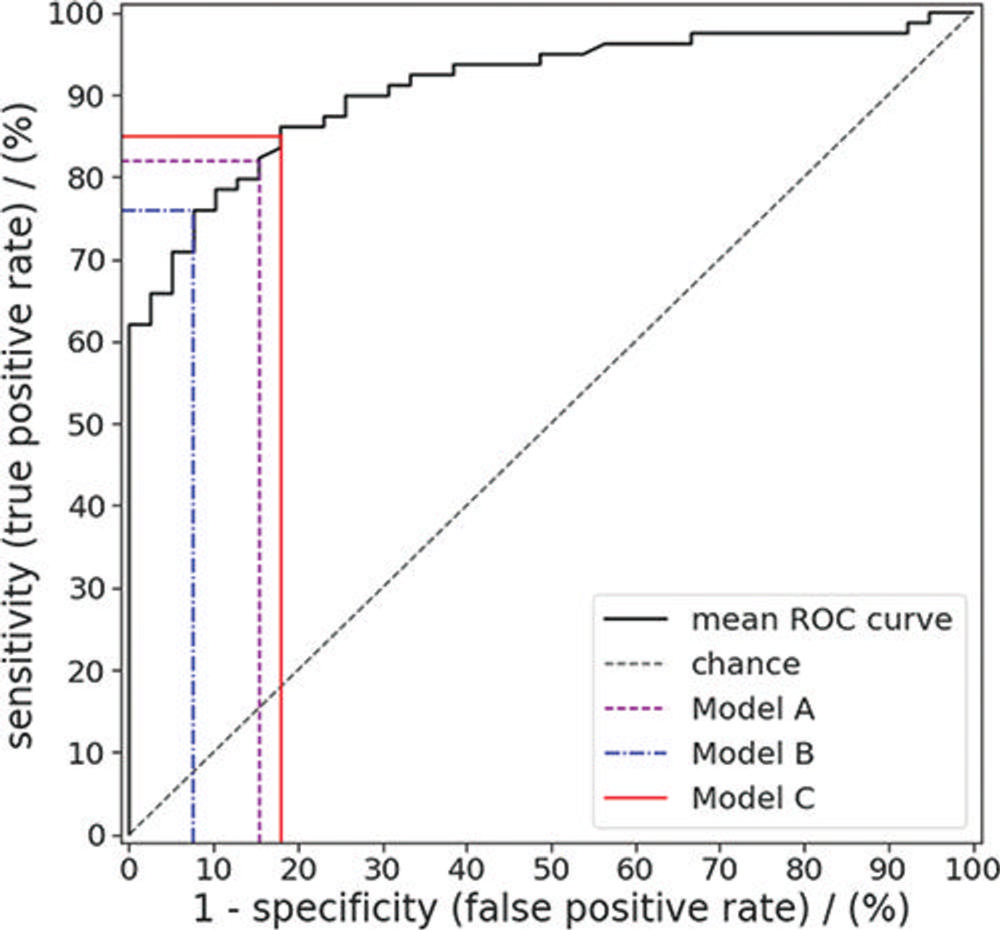
Figure 6. Receiver operating characteristic (ROC) curve for random forest predictions of colorectal polyp class (benign vs premalignant) for the polyps in the external test set. Sensitivity and specificity were evaluated for a default threshold value of 0.5 (model A), a threshold value that maximized the Youden index (model B), and a threshold resulting in the highest possible specificity while achieving a sensitivity of at least 85% (model C).
High-res (TIF) version
(Right-click and Save As)
