Artificial Intelligence Boosts MRI Detection of ADHD
Released: December 11, 2019
At A Glance
- Diagnosing ADHD is challenging, relying largely on symptoms and behavior-based tests.
- A deep learning model using multi-scale brain connectome data significantly improved MRI performance in detecting ADHD.
- The deep learning approach could also have applications for other neurological conditions.
- RSNA Media Relations
1-630-590-7762
media@rsna.org - Linda Brooks
1-630-590-7738
lbrooks@rsna.org - Dionna Arnold
1-630-590-7791
darnold@rsna.org
OAK BROOK, Ill. — Deep learning, a type of artificial intelligence, can boost the power of MRI in predicting attention deficit hyperactivity disorder (ADHD), according to a study published in Radiology: Artificial Intelligence. Researchers said the approach could also have applications for other neurological conditions.
The human brain is a complex set of networks. Advances in functional MRI, a type of imaging that measures brain activity by detecting changes in blood flow, have helped with the mapping of connections within and between brain networks. This comprehensive brain map is referred to as the connectome.
Increasingly, the connectome is regarded as key to understanding brain disorders like ADHD, a condition that makes it difficult for a person to pay attention and control restless behavior.
According to the National Survey of Children's Health, approximately 9.4% of U.S. children, ages 2 to 17 years (6.1 million) in 2016 have been diagnosed with ADHD. The disorder cannot yet be definitively diagnosed in an individual child with a single test or medical imaging exam. Instead, ADHD diagnosis is based on a series of symptoms and behavior-based tests.
Brain MRI has a potential role in diagnosis, as research suggests that ADHD results from some type of breakdown or disruption in the connectome. The connectome is constructed from spatial regions across the MR image known as parcellations. Brain parcellations can be defined based on anatomical criteria, functional criteria, or both. The brain can be studied at different scales based on different brain parcellations.
Prior studies have focused on the so-called single-scale approach, where the connectome is constructed based on only one parcellation. For the new study, researchers from the University of Cincinnati College of Medicine and Cincinnati Children's Hospital Medical Center took a more comprehensive view. They developed a multi-scale method, which used multiple connectome maps based on multiple parcellations.
To build the deep learning model, the researchers used data from the NeuroBureau ADHD-200 dataset. The model used the multi-scale brain connectome data from the project's 973 participants along with relevant personal characteristics, such as gender and IQ.
The multi-scale approach improved ADHD detection performance significantly over the use of a single-scale method.
"Our results emphasize the predictive power of the brain connectome," said study senior author Lili He, Ph.D., from the Cincinnati Children's Hospital Medical Center. "The constructed brain functional connectome that spans multiple scales provides supplementary information for the depicting of networks across the entire brain."
By improving diagnostic accuracy, deep-learning-aided MRI-based diagnosis could be critical in implementing early interventions for ADHD patients. Approximately 5% of American pre-school and school-aged children have been diagnosed with ADHD. These children and adolescents face a high risk of failing in academic study and building social relationships, which can result in financial hardship for families and create a tremendous burden on society.
The approach also has potential beyond ADHD, according to Dr. He.
"This model can be generalized to other neurological deficiencies," she said. "We already use it to predict cognitive deficiency in pre-term infants. We scan them soon after birth to predict neurodevelopmental outcomes at two years of age."
In the future, the researchers expect to see the deep learning model improve as it is exposed to larger neuroimaging datasets. They also hope to better understand the specific breakdowns or disruptions in the connectome identified by the model that are associated with ADHD.
"A Multichannel Deep Neural Network Model Analyzing Multiscale Functional Brain Connectome Data for Attention Deficit Hyperactivity Disorder Detection." Collaborating with Dr. He were Ming Chen, B.S., Hailong Li, Ph.D., Jinghua Wang, Ph.D., Jonathan R. Dillman, M.D., M.Sc., and Nehal A. Parikh, D.O., M.S.
Radiology: Artificial Intelligence is edited by Charles E. Kahn Jr., M.D., University of Pennsylvania (Penn) Perelman School of Medicine, Philadelphia, and owned and published by the Radiological Society of North America, Inc. (https://pubs.rsna.org/journal/ai)
RSNA is an association of over 53,400 radiologists, radiation oncologists, medical physicists and related scientists, promoting excellence in patient care and health care delivery through education, research and technologic innovation. The Society is based in Oak Brook, Ill. (RSNA.org)
For patient-friendly information on brain MRI, visit RadiologyInfo.org.
Images (.JPG and .TIF format)
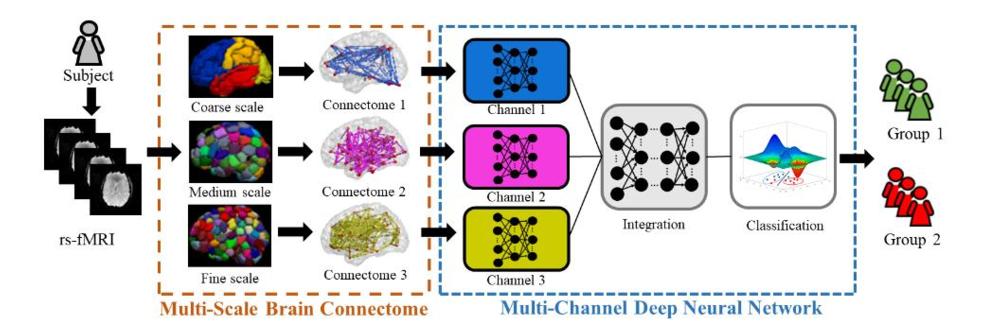
Figure 1. Schematic diagram of the proposed multichannel deep neural network model analyzing multiscale functional brain connectome for a classification task. rsfMRI = resting-state functional MRI.
High-res (TIF) version
(Right-click and Save As)
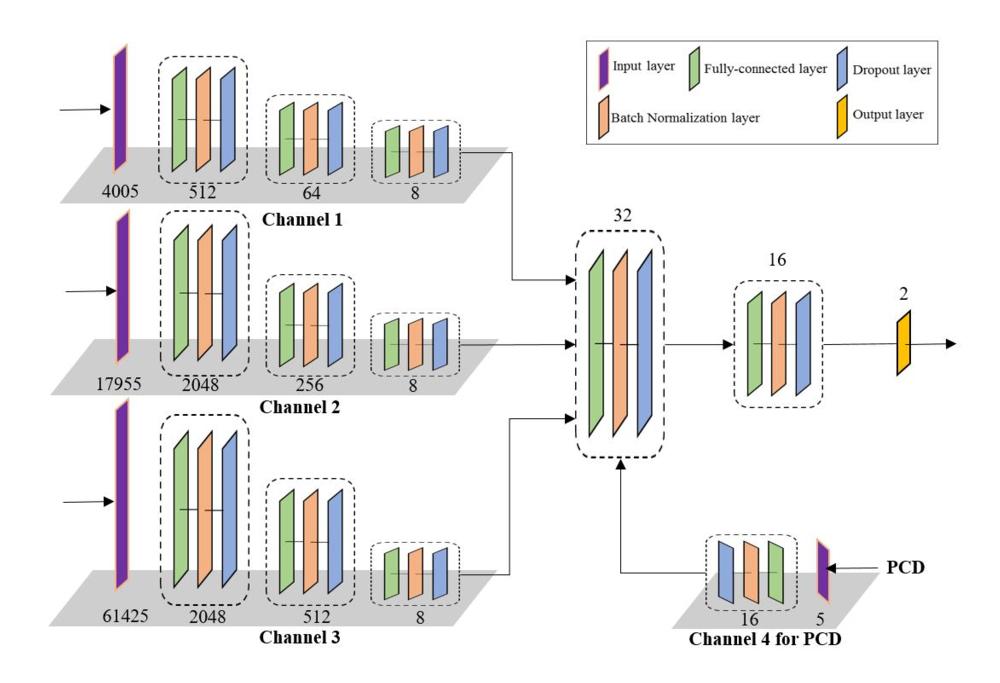
Figure 2. The architecture of a proposed multichannel deep neural network (mcDNN). The mcDNN has four input channels that take vectors of 1 X 7 for personal characteristic data and 1 X 4005, 1 X 17955, and 1 X 61425 for connectome data, consisting of the upper triangular values of the symmetric connectome matrices at a different scale. Four channels are eventually fused into one output channel. Each block reduces the feature dimensions. The number of feature dimensions is shown on the bottom of each block. PCD = personal characteristic data.
High-res (TIF) version
(Right-click and Save As)
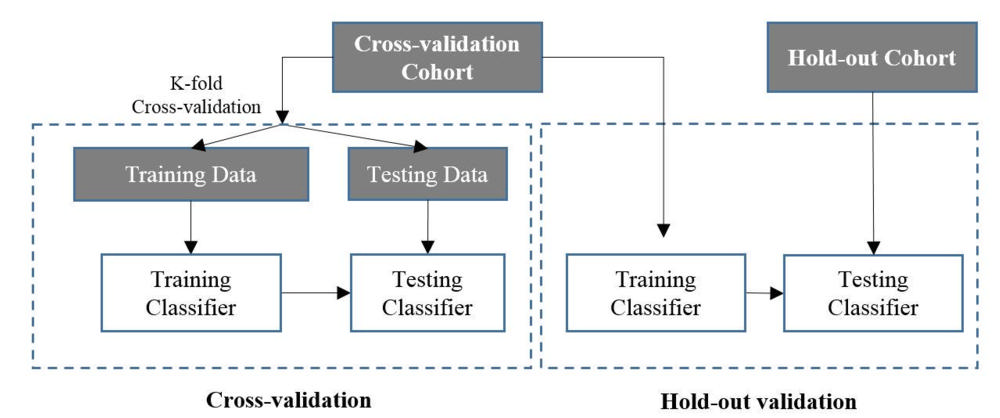
Figure 3. : Schemes of cross-validation and hold-out validation.
High-res (TIF) version
(Right-click and Save As)
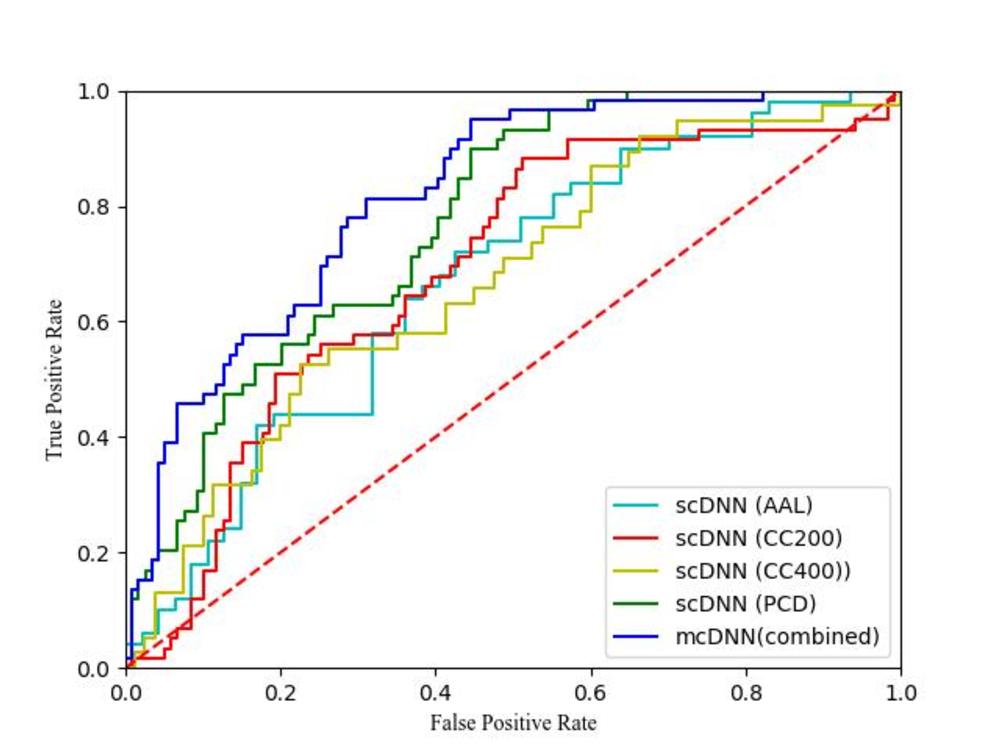
Figure 4. : Cross-validation. Receiver operating characteristic (ROC) curves of four single-channel deep neural networks (scDNNs) using brain connectome with parcellations of AAL, CC200, CC400, and PCD, independently, and our proposed multichannel deep neural network (mcDNN) using combined features (fusion of the multiscale brain connectome data and PCD). Areas under the ROC curves are 0.67, 0.69, 0.67, 0.77, and 0.82, respectively. Red dotted line signifies the "random guess". AAL = automated anatomic labeling, CC200, CC400 = brain connectome constructed on functionally defined parcellations, PCD = personal characteristic data (age, sex, handedness, and three individual measures of intelligence quotients).
High-res (TIF) version
(Right-click and Save As)
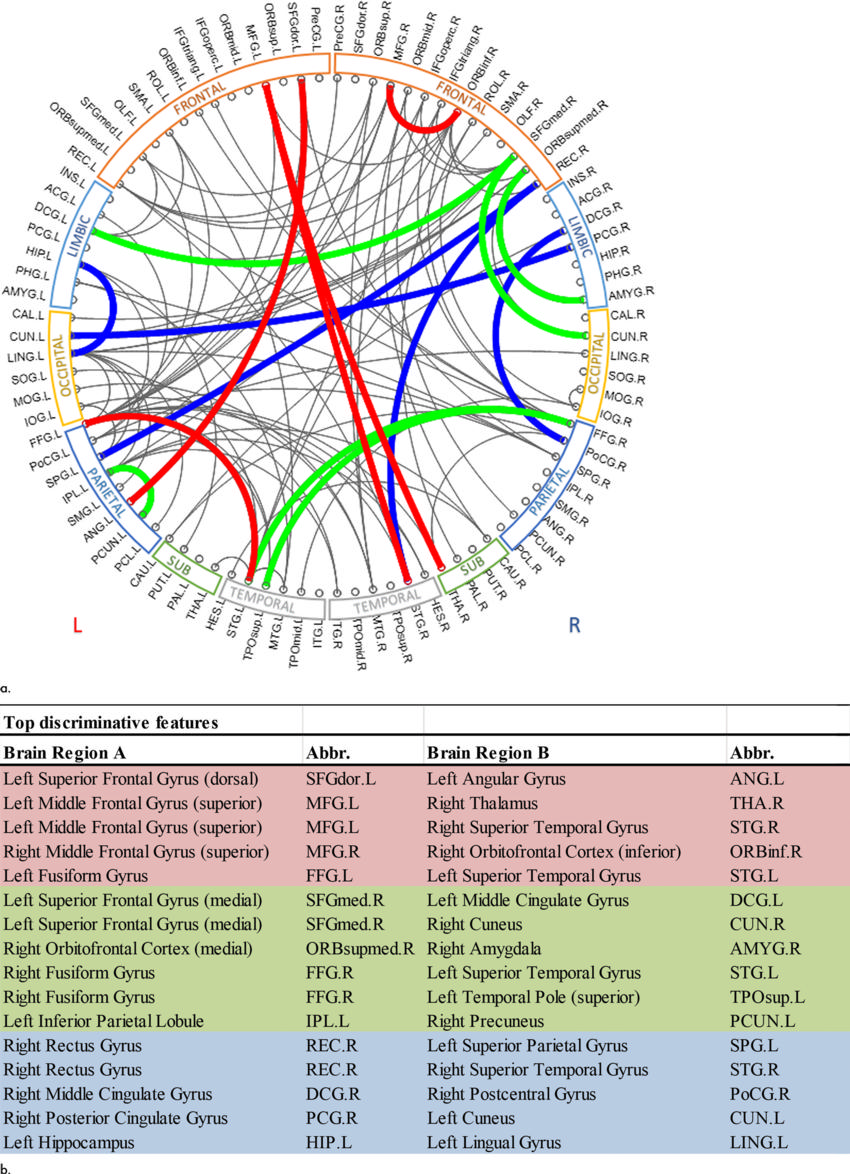
Figure 5. :Top discriminative functional connections. (a) By registering the atlases of CC200 and CC400 to AAL (mapping the spatial locations of top predictive connections of CC200 and CC400 onto the AAL), five common connections were identified at all scales (red), 11 common connections identified at the scale of AAL and CC200 (green and red); 10 common connections identified at the scale of AAL and CC400 (blue and red); and five common connections identified at the scale of CC200 and CC400 (red). (b) Table format shows top discriminative functional connections (from brain region A to brain region B) and their abbreviations. AAL = automated anatomic labeling, CC200, CC400 = brain connectome constructed on functionally defined parcellations, L = left, R = right.
High-res (TIF) version
(Right-click and Save As)
